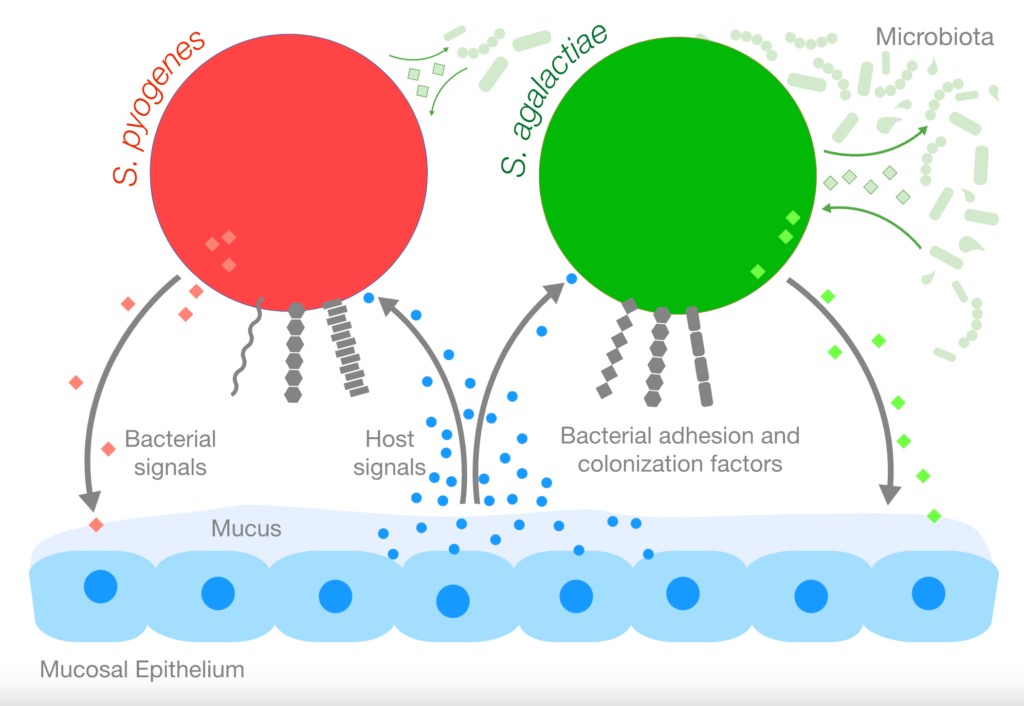
The Gram-positive bacteria Streptococcus pyogenes (Group A Strep, GAS) and Streptococcus agalactiae (Group B Strep, GBS) can cause invasive infections but are also able to colonize the host asymptomatically. The Cook lab focuses on determining the mechanisms behind how GAS and GBS colonize host mucosal surfaces.
Projects in the Cook lab involve examining the host factors that serve as signals to colonizing bacteria, the bacterial transcriptional changes elicited by these signals, and the phenotypic effects of these transcriptional alterations. We are also interested in the role of the normal microbiota in colonization and infection by GAS and GBS. The lab uses systems such as cell culture and mouse models of carriage and infection in addition to molecular and “-omics” approaches to examine interplay between the host and bacteria.